Comprehensive multi-omics analysis categorises distinct pathogenic processes in CLL
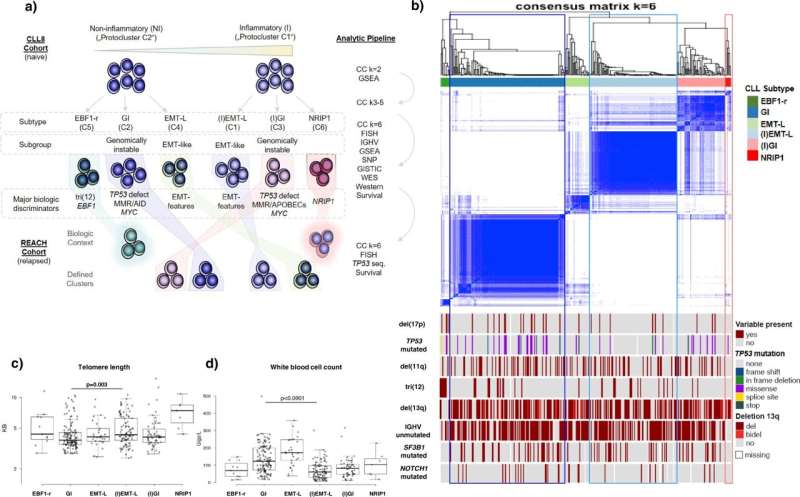
An international team of researchers has now comprehensively profiled and categorized over 700 tumor samples from patients with chronic lymphocytic leukemia (CLL) by analyzing multiple levels of encoded biologic information.
Through detailed mapping of the derived information, they identified major biologic categories associated with distinct modes of resistance to different treatment combinations.
Besides changes indicating heterogeneous levels of inflammatory activity, the degree of genomic instability turned out to be a major discriminatory feature for the tumor subgroups.
In cancer biology, high levels of genomic instability increase the likelihood for alterations in the genome and largely depend on the tumors capabilities to recognize and repair acquired DNA-damage.
Genomic instability in CLL has been linked to mutations or chromosomal losses of regions encoding the cellular watchers for genome protection, the so-called tumor suppressor genes.
Tumors exhibiting inactivation of tumor suppressor genes usually have a poor outcome and early detection helps to choose the most appropriate treatments. However, genomic instability also occurs in many CLL cases without such detectable alterations.
As reported by Johannes Bloehdorn (Ulm University) and colleagues in Nature Communications, tumor sample analysis included the assessment of gene mutations, chromosomal aberrations, DNA-methylation, gene and protein expression, and associated pathway-signaling activity.
The authors were able to decode the tumor biology and translate the underlying information in a way that makes it possible to categorize tumors irrespective of detectable losses or mutations of tumor suppressor or DNA-repair genes.
The authors further discovered that genomically instable tumors do not just shut down DNA repair but show boosted and highly error-prone repair activity. This further increases accumulation rates of genomic alterations, accelerating the development of resistance.
Another major subgroup identified in this study, showing the lowest degree of genomic instability, exhibits characteristics observed during the process of metastasis in solid tumors, which were previously not known to build subgroup-specific functional networks in CLL.
CLL cells can reach any part of the body via the bloodstream. Therefore, it appears redundant to develop mechanisms that enable the cell to break free from a tumor to search for a better survival niche, which is represented through the process of metastasis in solid tumors.
However, these uncovered networks may increase the ability for leukemia cells to exit the bloodstream and migrate to lymphoid tissues. Several targeted treatment approaches address related mechanisms and the cells ability to accumulate in lymphoid organs.
Source: Read Full Article