AlphaFold AI predicts SARS-CoV-2 omicron variant might not evade antibody neutralization
Scientists from the University of North Carolina have recently conducted computational modeling studies to predict the structure and immune escape ability of spike receptor-binding domain (RBD) of the recently identified Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The findings reveal that despite some mutations in the spike RBD, the Omicron variant is not capable of fully escaping the host immune responses.
The study is currently available on the bioRxiv* preprint server.
Study: Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions. Image Credit: Lightspring/Shutterstock
Background
The omicron variant of SARS-CoV-2 has been identified for the first time in South Africa in November 2021. The whole-genome sequencing analysis has revealed that the variant is heavily mutated, with 37 mutations in the spike protein and 15 mutations specifically in the spike RBD. On November 26, 2021, the World Health Organization (WHO) has designated the variant as a variant of concern (VOC). However, not enough evidence is so far available to understand the infectivity, transmissibility, pathogenicity, and immune escape ability of the Omicron variant.
In the current study, the scientists have computationally predicted the structure of the Omicron spike RBD from available sequences. Using the predicted structure, they have explored the dynamics of antigen-antibody interactions.
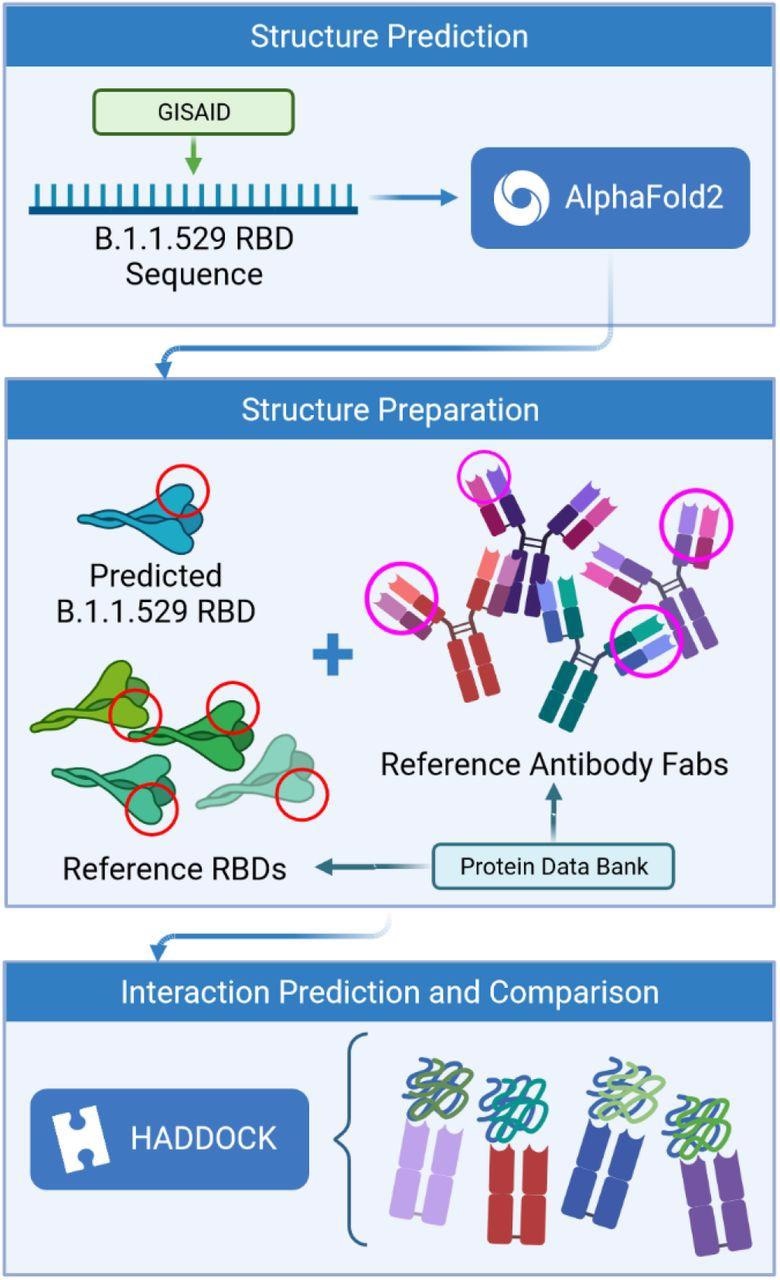
Study design
The scientists used the amino acid sequence of the Omicron RBD to generate a three-dimensional structure via a deep learning model. Afterward, they simulated the interaction of predicted RBD structure with available structures of four neutralizing antibodies. The structures of these antibodies were obtained from SARS-CoV-2-infected patients.
They used the AlphaFold is an artificial intelligence program to predict the binding affinity of antigen-antibody interactions. They assessed interactions between epitope (antibody binding site) and paratope (antigen-binding site) structures to finally predict whether mutations in the RBD can facilitate the virus escape antibody-mediated neutralization.
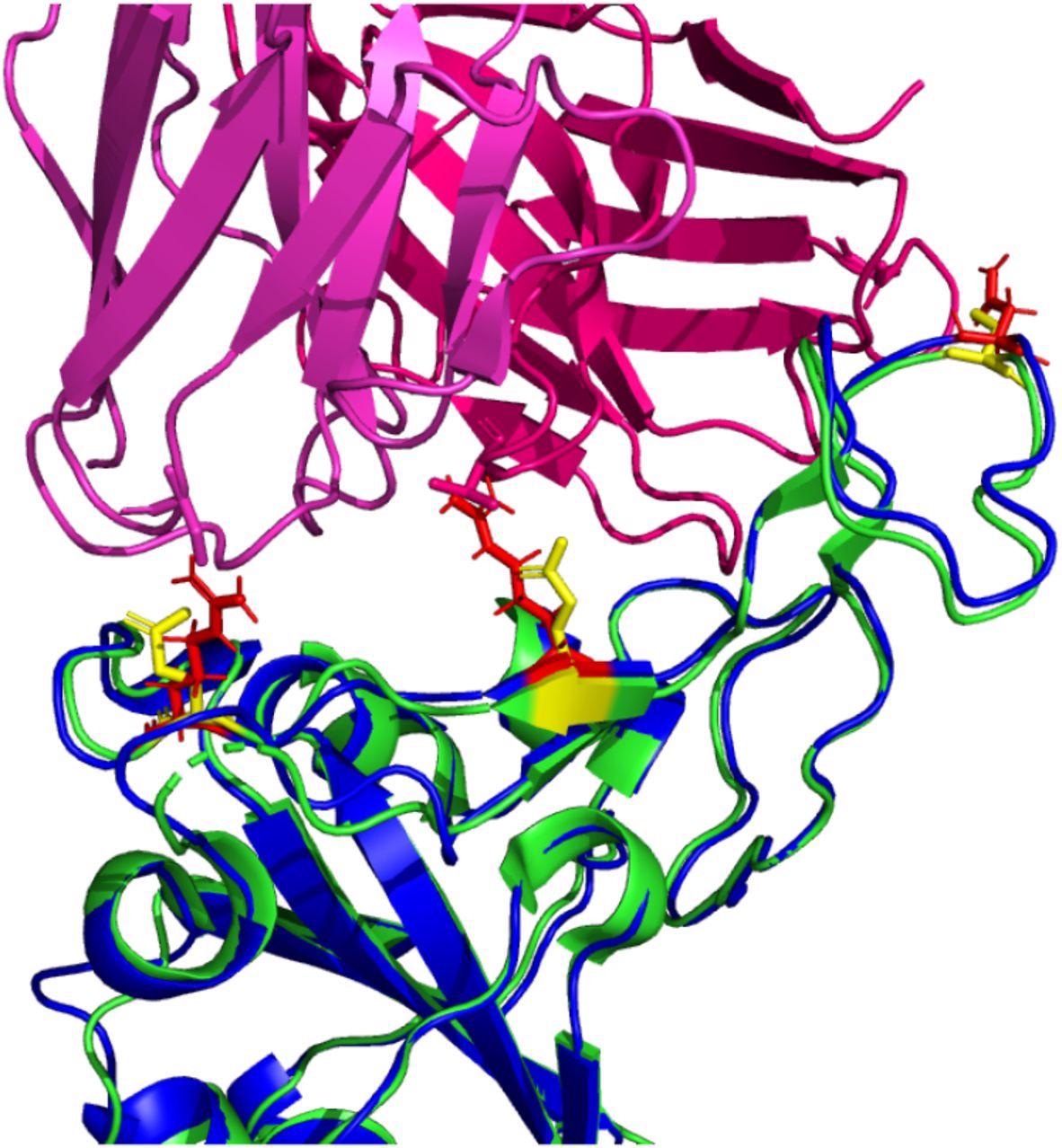
Important observations
The receptor-binding motif (RBM) of the Omicron variant contains nine characteristic mutations. The analysis of variant-specific sequences revealed that the Omicron variant is distantly related to the original Wuhan strain of SARS-CoV-2 and its Gamma variant (first identified in Brazil).
The mutational analysis revealed that the Omicron RBD contains 15 single amino acid substitution mutations in the RBD that resulted in alterations in residue types. These mutations had caused changes in amino acid polarity and length of side-chains. These changes are expected to alter the binding affinity between RBD and antibody by either changing the surface charges or by creating steric hindrance.
Antigen-antibody interactions
The antibody binding analysis revealed that the omicron RBD has a lower binding affinity for each tested antibody compared to the wild-type RBD. There was a 4% – 52% reduction in electrostatic interaction. Moreover, the findings revealed a reduction in buried surface area, indicating that the omicron RBD interacts with the antibodies more distantly. Importantly, the statistical analysis revealed that the binding affinity of omicron RBD for tested antibodies is not significantly different from that of wild-type RBD.
The analysis of omicron RBD/wild-type RBD – antibody complexes highlighted the changes in certain residues, including 448N, 484A, and 494S, at the binding interface. In contrast, certain residues remained unchanged in the Omicron RBD – antibody binding interface. Overall, the analysis revealed that the majority of RBD mutations in the Omicron variant do not cause significant conformational changes that can severely affect the binding ability of neutralizing antibodies.
Study significance
The study utilizes in silico predictive tools to determine the structure of omicron RBD and its interaction dynamics with anti-SARS-CoV-2 neutralizing antibodies. As observed in the study, despite acquiring 60 mutations in the RBD, the omicron variant is not able to completely escape antibody-mediated neutralization. The reduction in binding affinity observed between omicron RBD and tested antibodies could be because of the substitutions to amino acids with larger side-chains. These substitutions in the omicron RBD have resulted in slightly more distant interaction with antibodies, and thus, have caused a reduction in binding affinity compared to wild-type RBD.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Ford CT. 2021. Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions. bioRxiv. doi: https://doi.org/10.1101/2021.12.03.471024 https://www.biorxiv.org/content/10.1101/2021.12.03.471024v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Amino Acid, Antibodies, Antibody, Antigen, Artificial Intelligence, binding affinity, Computational Modeling, Coronavirus, Coronavirus Disease COVID-19, Deep Learning, Genome, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article