Mosaic-8 RBD-nanoparticles show promise against current and future SARS-CoV-2 variants
A recent study posted to the bioRxiv* preprint server supports using mosaic sarbecovirus receptor-binding domain (RBD)-nanoparticles to discover therapeutic pan-variant and pan-sarbecovirus monoclonal antibodies (mAbs) and to induce mAbs by vaccination.
Background
Analysis of the variability of RBDs between sarbecoviruses and inside severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of interest (VOIs) and variants of concern (VOCs) suggests that vaccines and mAbs addressing the highly conserved neutralizing antibody epitopes, i.e., class 4 and class 1/4, can safeguard from the current and future SARS-CoV-2 VOCs and stop forthcoming sarbecovirus spillover incidents from causing another pandemic or epidemic.
The current study's authors previously illustrated a vaccine strategy involving the parallel display of sarbecovirus RBDs on protein-based nanoparticles. This approach demonstrated improved heterologous adhesion, neutralization, and immunity from sarbecovirus challenges in animal models compared with homotypic, i.e., SARS-CoV-2 RBD-only, nanoparticles.
About the study
In the present investigation, the researchers from Beckman Research Institute of City of Hope, the California Institute of Technology and Stanford University evaluated the RBD epitopes of mAbs derived from mosaic (SARS-CoV-2+seven animal sarbecovirus spike (S)) RBD- and homotypic (i.e., SARS-CoV-2-only) RBD-immunized mice to profile the antibody reactions to RBD nanoparticles.
The team developed multimerized RBD-nanoparticles by covalently attaching RBDs to C-terminal SpyTag003 sequences employing the SpyCatcher-SpyTag platform to a 60-mer nanoparticle, i.e., SpyCatcher003-mi3. They established and described nanoparticles containing randomly ordered RBDs from seven animal sarbecovirus and SARS-CoV-2 Ss referred to as mosaic-8 RBD-mi3, as well as nanoparticles comprising solely SARS-CoV-2 RBDs, known as homotypic SARS-CoV-2 RBD-mi3.
The authors first examined the adhesion of 13 purified mAb immunoglobulin Gs (IgGs) to SARS-CoV-2 variants and other sarbecoviruses RBDs employing enzyme-linked immunosorbent (ELISAs) tests. They analyzed the mAb adhesion characteristics to four human anti-RBD IgGs with established epitopes: C118, sotrovimab (S309), C102, and C144. They conducted subsequent assessments using seven mAbs obtained from IgG-secreting B cells coupled to two or more RBDs in screening.
Using a pseudovirus neutralization evaluation, the investigators assessed the mAbs neutralization capacities of sarbecoviruses leveraging human angiotensin-converting enzyme 2 (ACE2) for target cell entrance. These sarbecoviruses include SARS-CoV-2 Beta, WA1 D614G, Omicron BA.2 and BA.1 and Delta variants, SARS-CoV, SHC014, WIV1, a modified LyRa3/SARS-CoV Chimera, and BtKY72 Khosta2/SARS-CoV Chimera. They conducted a binding experiment to determine possible competition with proteins that attach to recognized RBD epitopes to find RBD epitopes targeted by the mAbs.
The team conducted structural investigations of the seven mAbs shown to bind two or more RBDs in Beacon isolation to determine recognition and neutralizing processes. Besides, the contacts of HSW-2 mAb (derived from homotypic SARS-CoV-2 RBD nanoparticle-immunized mice) with the S trimer were examined using single-particle cryogenic-electron microscopy (cryo-EM).
The authors examined the fragment antigen-binding (Fab)-bound S trimer structures they found with other trimer structures to explore the potential impact of mAb attachment with S trimer conformation. The scientists plotted the adhesion epitopes of the mAbs for which they obtained Fab-S configurations relative to the positions of Omicron BA.2 and BA.1 alterations on the RBD to delineate how alterations in recent VOCs would affect the binding of those mAbs. Furthermore, they used modeling to determine how the RBD-nanoparticles employed to elicit the mAbs currently assessed would interact with bivalent B cell receptors.
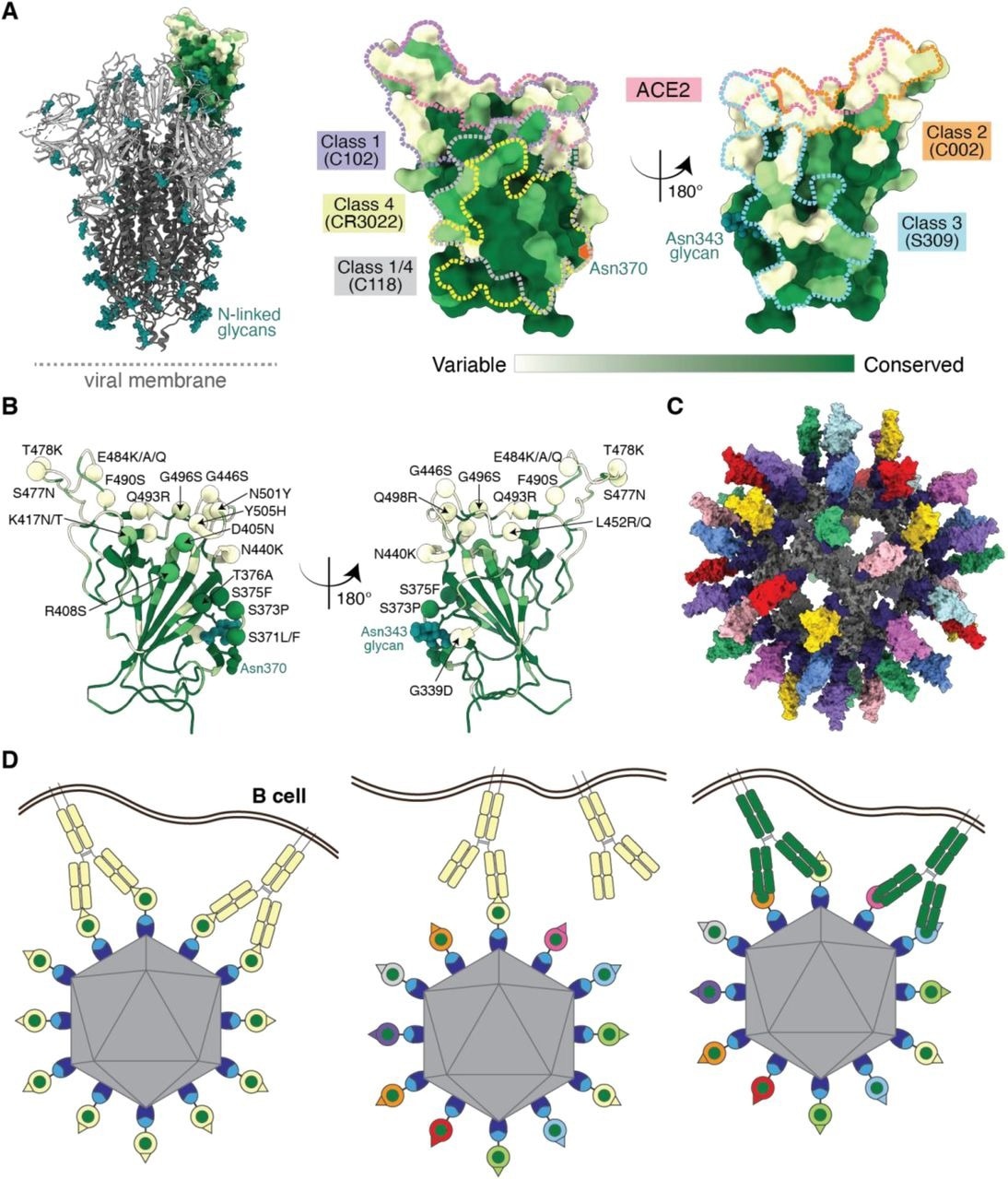
Utilizing antibody avidity effects suggests a strategy to target antibodies to conserved regions of sarbecovirus RBDs. (A) Left: Structure of SARS-CoV-2 spike trimer (PDB 6VYB) with one RBD in an “up” position. Right: Sequence conservation of 16 sarbecovirus RBDs (Figure S1) calculated by the ConSurf Database (Landau et al., 2005) plotted on a surface representation of the RBD structure (PDB 7BZ5). Class 1, 2, 3, 4, and 1/4 epitopes are outlined in different colored dots using information from structures of the representative monoclonal antibodies bound to RBD or spike trimer (C102: PDB 7K8M; C002: PDB 7K8T, S309: PDB 7JX3; CR3022: PDB 7LOP; C118: PDB 7RKV). (B) RBD mutations of 13 SARS-CoV-2 VOCs and VOIs (https://viralzone.expasy.org/9556) plotted onto the RBD structure (PDB 7BZ5) as spheres that are colored according to the variability gradient in panel A. The N-linked glycan at position 343 of SARS-CoV-2 RBD is shown as teal spheres, and a potential N-linked glycosylation site at position 370 (SARS-CoV-2 numbering) found in some sarbecovirus RBDs but not in the SARS-CoV-2 RBD is indicated by an orange hexagon. (C) Structural model of mosaic-8 nanoparticle formed by SpyCatcher-mi3 and eight SpyTagged RBDs made using coordinates of an RBD (PDB 7SC1), mi3 (PDB 7B3Y), and SpyCatcher (PDB 4MLI). (D) Hypothesis for preferential stimulation of B cells with that encode cross-reactive antibodies by mosaic (right) versus homotypic (left) RBD nanoparticles. Left: Yellow B cell receptors recognizing an accessible strain-specific epitope (yellow triangle) can crosslink between adjacent RBDs on a homotypic nanoparticle to enhance binding through avidity effects. Middle: Yellow B cell receptors against a strain-specific orange epitope cannot crosslink between adjacent RBDs on a mosaic RBD nanoparticle that presents different versions of the epitope (colored triangles). Right: Green cross-reactive B cell receptors can crosslink between a conserved epitope (green circles) on adjacent RBDs in a mosaic RBD nanoparticle to enhance binding to a more occluded, but conserved, epitope through avidity effects.
Results and conclusions
The study results demonstrated that mosaic sarbecovirus RBD-nanoparticles triggered robust neutralizing antibodies that cross-react among SARS-CoV-2 VOCs and animal sarbecoviruses.
Although the team discovered just five mAbs from mosaic-8 nanoparticle-immunized mice that coupled to two or more RBDs, one mAb, i.e., M8a-3, was potently neutralizing and cross-reactive. Moreover, two others, i.e., M8a-34 and M8a-31, were less strongly neutralizing yet were cross-reactive. Additionally, another mAb, i.e., M8a-28, robustly neutralized SARS-CoV-2 variants and was similar to therapeutic antibodies in present use, e.g., Bebtelovimab. These findings illustrated the capacity of the mosaic-8 immunogen to effectively induce possibly beneficial therapeutic antibodies that could be recognized in extensive assessments and established as future therapeutic mAbs.
Structural analyses of Fab-SARS-CoV-2 S trimer complexes, such as one with Omicron BA.1, indicated that four out of five mAbs derived from mosaic-8 immunized mice identified conserved epitopes, as devised in the immunization strategy and as demonstrated for polyclonal antisera generated in mice via mosaic-8 RBD-nanoparticle immunization. Contrarily, antibodies from mice immunized with homotypic SARS-CoV-2 RBD-nanoparticle more frequently identified variable class 2 and 1 RBD epitopes. This inference probably explains why it was more challenging in the present study to extract single B cells from homotypic SARS-CoV-2 RBD-nanoparticle-immunized mice (HSW) that secreted IgGs coupled with two or more labeled RBDs.
The two cross-RBD attaching mAbs that the researchers isolated from mice immunized with homotypic RBD-nanoparticle exhibited adherence to numerous sarbecovirus RBDs yet were either poorly or non-neutralizing. Structural analyses explained the HSW mAbs' poor neutralization characteristics. In contrast to the structures of more powerfully neutralizing mAbs, which revealed three attached Fabs for each trimer, the HSW-1-S configuration only displayed one attached Fab per trimer. Further, a trimer separation and a Fab-monomeric S1 subunit structure resulted from the HSW-2 Fab epitope's inability to bind to its RBD epitope on an intact S trimer.
Altogether, the current findings, combined with the prior reports, reinforce the mosaic-8 RBD-nanoparticles as a potential vaccine option to safeguard from prospective spillover sarbecoviruses from animal hosts and SARS-CoV-2, including future variants. Additionally, since efficient cross-reactive mAbs were recognized from fewer B cells, a high-throughput assessment of large-scale samples from mosaic-8 RBD-mi3-immunized animals could be employed in identifying diverse clinical mAbs. These mAbs then might assist in treating or preventing SARS-CoV-2 Omicron or future variant infections.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Neutralizing monoclonal antibodies elicited by mosaic RBD nanoparticles bind conserved sarbecovirus epitopes; Chengcheng Fan, Alexander A Cohen, Miso Park, Alfur Fu-Hsin Hung, Jennifer R Keeffe, Priyanthi NP Gnanapragasam, Yu E Lee, Leesa M Kakutani, Ziyan Wu, Kathryn E Malecek, John C Williams, Pamela J Bjorkman. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.06.28.497989, https://www.biorxiv.org/content/10.1101/2022.06.28.497989v1
Posted in: Molecular & Structural Biology | Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Antigen, B Cell, Cell, Coronavirus, Coronavirus Disease COVID-19, Electron, Electron Microscopy, Enzyme, Glycan, Glycosylation, immunity, Immunization, Immunoglobulin, Microscopy, Nanoparticle, Nanoparticles, Omicron, Pandemic, Protein, Pseudovirus, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Sotrovimab, Syndrome, Vaccine

Written by
Shanet Susan Alex
Shanet Susan Alex, a medical writer, based in Kerala, India, is a Doctor of Pharmacy graduate from Kerala University of Health Sciences. Her academic background is in clinical pharmacy and research, and she is passionate about medical writing. Shanet has published papers in the International Journal of Medical Science and Current Research (IJMSCR), the International Journal of Pharmacy (IJP), and the International Journal of Medical Science and Applied Research (IJMSAR). Apart from work, she enjoys listening to music and watching movies.
Source: Read Full Article